Information for Astronomers
User Tools
Sidebar
Table of Contents
Reduction of pointing measurements
Location of the Raw Data
The raw data of the 100m Effelsberg telescope is stored in MBFITS-Format. In Effelsberg the files are located in the directory /daten/Raw which should be available on every Observer-PC. Older data can be found in /eff/data/Raw/Raw-YYYY-MM. /eff/data/Raw is available on every Observer-PC and in the network in Bonn as well. Most programs listed here support the flag "fdir" to point the program to the directory where the MBFITS data can be found. Default is always /daten/Raw.
Every 30 minutes the raw data is synced to Bonn. It is accessible in /eff/data/. Old data is accessible in /hsm/effarchive More details on the location of data can be found here under Data storage and archive
Current Observer-PCs are observer4 (64-bit) and observer7 (32-bit). Observers should use their MPIfR account to login to those machines, observes without an MPIfR account can use the common obs2 account.
Inspecting scans by hand using the Toolbox
The MBFITS data can be inspected with any program that understand FITS Format e.g. "fv" fits-viewer. You can look at the headers and tables and plot different data columns…
However, most users might prefer a kind of pre-reduced view where you see the amplitude of the scan calibrated in units of the calibration temperature and with real arcseconds for the scanning axis. This is provided by the "toolbox" program. The toolbox can be used on the observer-PCs (currently observer7 and observer4) using your normal MPI account. You only need to add /opt/bin to your PATH variable.
#bash export PATH=$PATH:/opt/bin #csh set path = ($path /opt/bin)
Observers that don't have a MPI user Account can use the common obs2 account.
The toolbox can be used interactively by calling
toolbox
from the command line. A file browser opens from which the scan can be selected (see Fig. 1). You can also select whether you want to have a ps-plot of the scan written to your current directory. By default you will find a .fit-file for every scan you looked at with some scan specific data and the fitted parameters.
Figure 1 Toolbox file browser.
Content of fit-file:
# # Source Scan direct Subsc MJD LST Azi Elv Amp err Offset err FWHM err ColS NulE Parangle Tsys Cal 3C286 4047 ALON 1 55497.2278085 30085.1592055 78.008 30.621 7.61715e-01 6.32183e-03 9.59 0.42 69.78 0.84 16.51 7.39 -46.17 8.673e+00 1.8370e+03 3C286 4047 ALON 2 55497.2281492 30114.6799258 78.097 30.697 7.66583e-01 5.80441e-03 -0.38 0.38 68.84 0.75 16.51 7.39 -46.19 8.668e+00 1.8373e+03 3C286 4047 ALAT 3 55497.2285233 30147.0884189 78.188 30.780 7.62980e-01 8.15973e-03 0.82 0.54 69.42 1.07 16.51 7.39 -46.21 8.659e+00 1.8389e+03 3C286 4047 ALAT 4 55497.2288663 30176.8003259 78.274 30.859 7.61237e-01 7.00963e-03 1.35 0.46 68.80 0.91 16.51 7.39 -46.23 8.652e+00 1.8386e+03
Another option, that is more appropriate for reducing a number of scans, is to use the toolbox from the command line:
toolbox scan=XXXX plot='/xs'
This will reduce scan XXXX and shows the plot in a PGPlot window (see Fig. 2).
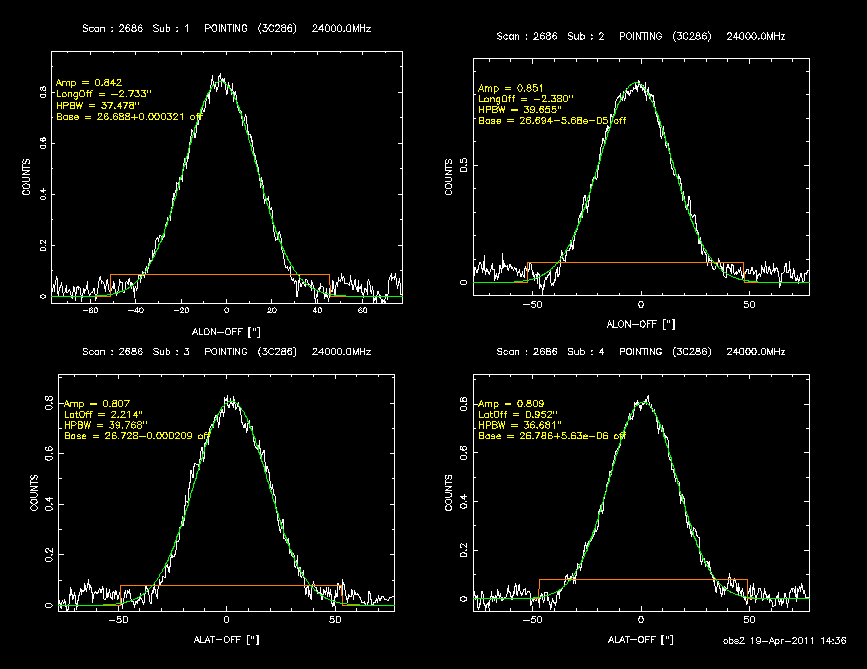
Figure 2 PGPlot window of a cross scan. The data is baseline subtracted and a Gaussian is fitted.
Toolbox Options
There are a number of options to control the toolbox. The order of their appearance on the command line is not important. Every keyword will be recognized.
| Option | Comment |
|---|---|
| fdir='PATH' | Path to the MBFITS data |
| scan=xxxx | Direct selection of scan numbers. No File-Browser will be opened. |
| scan=last | Will be open the most recent scan. |
| sub=X | Displays sub scans X only., e.g. sub=2 shows sub-scan 2 only. |
| take=XXXX:x1,x2,x3,YYYY:y1,y2,y3,… | Reduces the given sub scan numbers of scan XXXX and YYYY together. All scans have to be specified with scan=XXXX,YYYY. Works also together with "aver" (see below) |
| del=XXXX:x1,x2,x3,YYYY:y1,y2,y3,… | Excludes the given sub scan numbers from the reduction of scan XXXX and YYYY. All scans have to be specified with scan=XXXX,YYYY. Works also together with "aver" (see below) |
| plot | Enables postscript plots. Name of the plot: xxxx.ps, xxxx="scan number" |
| plot='/xs' or '/gif' | Opens plots in a PGPlot-XServer or writes gif-images: xxxx.gif |
| pol=u or q | in combination with plot the graphs for the Q and U stokes parameters will be shown |
| log10 | plots the y-axis in log10 scale |
| yscale=min,max | allows to restrict the plotted y-axis range, e.g., yscale=0,10 for numbers from 0 to 10. |
| nos5 | to reduce polarimetry data from receivers without the phase-switch (S5). |
| nofit | Displays the data as taken. No baseline subtraction and fitting is done. |
| baseline=[no,0,1, or 2] | Set the degree of the base line that is subtracted. no=no baseline, 0=constant offset, 1=liner fit, and 2=2nd degree polynomial. |
| cut=-x,x | Restricts the range of the Gaussian fit in scanning direction on the left (-x) and right (x). E.g. cut=0.1,0.1 will reduce the range by 10%. |
| aver | Causes an average over all longitude and latitude subscans |
| aver and scan=XXXX,YYYY,ZZZZ | Causes an average over all longitude and latitude subscans and scans (XXXX, YYYY, and ZZZZ). The resulting parameters will be written in XXXX+YYYY+ZZZZ.fit |
| use='(c[1]+c[2])/2' or '(fac1*c[1])+(fac2*c[2])/2' | Enable to add or subtract (e.g. for beam switch) any combination of available channels and to multiply them by factors. Useful to apply T_{cal} [K] to the measured counts to get T_A [K]. Which signals are exactly available is listed in the receiver folders in the control room and can be seen in the FEBEPAR header of the MBFITS file. As obs2 on Observer3 also FEBEinfo.py can be used to obtain information about the written channels. |
| spikes=X | Option to despike data with interference. X=hight of spike in multiples of the RMS (default 3). Despike is turned off if X=-1. |
| Writes out the data in ascii format. One file for every subscan with scan position, counts, and Gauss-fit is written. | |
| fcent=x | Defines the starting point (centre) of the Gaussian fitting, 'x' in arcsec. |
| febe=XXX | Sets the frontend/backend combination, XXX, for scans that are observed with more than one. |
| fit=n | Number of iterations for the Gaussian fitting. Default is 2. |
| header | Prints the MBFIT header. |
| horn=n | Defines the horn number 'n' to reduce. See also the "use" option, that is more flexible. |
| rfi=n | Will filter RFI peaks looking for peak increasing n times the rms. |
| window=n | Defines the window size in multiples of the beam width for the second iteration of the Gaussian fit (orange box in the plot window). Default is 1.22 |
| rms | Prints the rms of the scan after subtracting the fit. |
| mapping options: | For viewing maps or converting them to FITS or nod2 standards. |
| color | Shows color plots instead of black and white. |
| resize | Allows to plot the map on a new grid. |
| rfi=n | Will filter RFI peaks looking for peak increasing n times the rms. |
| tab=n | Allows to fill every n'th line with dummies for under sampled maps. |
| taper | Gaussian taper for the sinc-function. |
| xstart=n | Defines the start coordinate for the x-axis in degree. |
| more dangerous option: | "dangerous" means you have to know what you do |
| sig="(1,1)+(1,2)+(1,3)+(1,4)" | Definition of an individual formula to add channels and phases. Usually the phases 3 and 4 contain the calibration signal. |
| cal="-(1,1)-(1,2)+(1,3)+(1,4)" | Corresponding calibration formula. Phases 1 and 2 are subtracted to only get the calibration signal. |
| sig="(1,1)+(1,2)+(1,3)+(1,4)+(2,1)+(2,2)+(2,3)+(2,4)" | Another example: formula to add channel 1 and 2, e.g. LCP + RCP. But it is recommended to use the "use" option add or subtract channels. |
Observing Logs
All observations are logged in a MySQL data base. To see the content of the data base, call the program
obslog
on observer2. For more information see Obslogger.
Once you have found your observations in the Obslogger, the log can be saved in a text file that can be used as an input file for the further data reduction below.
Reducing a number of scans at once
Looking at single scan might be appropriate for checking the data during an observation, but for calibration or flux density monitoring a more automatic way is preferred. There is a collection of scripts and programs that can be used to perform all the tasks to obtain flux density calibrated data. The scripts are located in /opt/bin on the Observer-PCs.
Raw Data Processing
The scripts and programs mostly use a file that contains all the scan numbers to be reduced. The scripts dblog2scan.py or dblog2scan_wea.py produce such lists from observing logs written Obslogger.
In dblog2scan.py one can optionally restrict the frequency by the receiver version and frequency in GHz, if the log contains entries at different frequencies.
ubach@observer4:~$ dblog2scan_wea.py Usage: dblog2scan.py log-file [receiver] [freq GHz] Give a frontend designation (e.g. 28.1) and additional a frequency
dblog2scan.py will just produce the files scans (input file for weather.py and corr_point.py) and scanlist (to be attached to the reduce.par file), dblog2scan_wea.py will produce an additional weather.dat file with weather information for each scan. The weather file is useful for the opacity correction described below.
The script reduce.py can be used to reduce a number of given scans using the toolbox with a list of options. Some example parameter files are stored in /home/obs2/flux_monit/reduce-par. E.g., reduc28.par for the 2.8cm SFK receiver. Calling just reduce.py prints out some help as well.
obs2@observer9:~$ reduce.py Usage: reduce.py par-file The par-file consists of two parts 1. The Toolbox options: e.g. # Toolbox options start here start_options # # Beam switch use='(c[1]+c[2]-c[5]-c[6])/2' # # Use XServer from PGPlot plot='/xs' # # Average scans in ALON and ALAT aver end_options 2. A scan list: # # sub-scans can be deleted by the del= option # in the second line: e.g. # scan=0001 # del='1,4,5' # to delete sub-scans 1, 4, and 5 # # New root dir for data can be specified as e.g. # fdir=/daten/Raw/Raw-2011-01 # default is /daten/Raw scan=0001 scan=0002 scan=0003
For example:
> reduce.py reduc28.par Remove previous fit-files Start reducing data: toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8464 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8465 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8479 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8480 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8494 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8495 toolbox use='(c[1]+c[2])/2' plot='/xs' scan=8511 ...
will average over channel 1 and 2 and plot the scans in a PGPlot window. If you prefer to save the figures instead of looking at them as they are being processed try just "plot" or "plot='/gif'". All options from the table can be specified in options section of the parameter file. In the scan section each scan entry can be followed by a delete line to exclude some subscans from the data reduction. That might help in case of RFI or short term weather effects. The keyword "fdir" can be used to change the PATH to the data. It has to be given only once and the following scan numbers will be all processed from this directory.
The resulting fit-files have an entry for the system temperature. This can be used to correct the data for opacity effects (see next section). Since Tsys is calculated from the baseline parameters it only works when a baseline was fitted and it was done in total power. If one uses a software beam switch the baseline will just be the residuals between the two horns and does not correspond to Tsys any more. In the case you want to use the software beam-switch to improve the fitting of the Gaussian it might be wise to first do one run with total power to produce a Tsys-file and then run the procedure again using software beam-switch to obtain a better fit.
Correct for Opacity and Pointing Offset
A short introduction to the background of the further steps of the data calibration can be found here. The procedure described here that fits the lower envelope of the Tsys vs. airmass distribution only works if you have several scans covering a larger range of elevations. An alternative would be to perform skydips during the observations to measure the opacity and and lower frequencies one can also use the common values from our receiver page.
Since the amplitudes from the toolbox are in units of the Tcal one just have to multiply the given numbers by the Tcal to get the correct system temperature. The reduce.py procedure provides an all.fit file that contains all the single fit-files. The further programs work with this all.fit.
The script weather.py reads the weather information from the weather.dat file or directly from the MBFITS files. It multiplies the Tsys found in the all.fit by a given Tcal and prints out a LIST.tsys, with the opacity information for each scan that is listed in the scanlist-file. When the weather data is read from the raw MBFITS file the script will write a new weather.dat for alter use. The minimum zenith opacity is fitted from the Tsys vs. airmass distribution and is presented in the plot Opacities.eps. Weather.eps contains the weather info.
obs2@observer9:~$ weather.py Task to read weather data from MBFITS files, save them in ASCII format, and compute LIST.tsys. Usage: weather.py scan-file [Tcal] [fdir=<PATH>] The scan-file should contain the scan numbers to be reduced. Comments can be inserted with a leading '#', e.g.: # Good pointing scans 2567 2568 #2569 removed because of RFI! 2570 ... Options: Tcal: temperature of the noise diode to get correct Tsys (default=1) fdir=<PATH>: if the data are not in /daten/Raw any more
The script corr_point.py allows to apply the opacity corrections from LIST.tsys or to apply a single value for all data. The question how to proceed will be ask when the the script is started. The amplitudes will be corrected for pointing offsets. Offsets in longitude will be applied to the latitude data and vice versa. To calculate the correction the actual FWHM of the Gaussian fit will be used. If you don't trust that fit the value after the Tcal is interpreted as the FWHM to use.
obs2@observer9:~$ corr_point.py Task to reduce fit-files from Peters Toolbox Usage: corr_point.py scan-file [Tcal (K)] [FWHM (asec)] The scan-file should contain the scan numbers to be reduced. Comments can be inserted with a leading '#', e.g.: # Good pointing scans 2567 2568 #2569 removed because of RFI! 2570 ...
For example the Tcal at 2.8cm is 7.5 K and scan numbers are stored in the file scans.
obs2@observer3:~/ubach/measurements/2011_03_16_poi/2.8cm/0.Raw$ corr_point.py scans 7.5 **************************************************** * Procedure to analyse Cross-scans * * from the 100m Effelsberg telescope * * * * Version 3.4 (I. Marti-Vidal) * * (T. Krichbaum) * * (U. Bach) * * * * adapted from A. Kraus corr_point.f Ver. 2.7 * * * **************************************************** Raw data must be in file: all.fit Apply opacity correction? No -> [n] Apply a single value -> [s] Apply computed values (LIST.tsys) -> [f] n No opacity correction! Tcal=7.50 [K] Write LIST.raw Write Pointing.dat In total there have been 312 data sets in 156 Scans. <FWHM> = 69.09 +- 1.42" <Off_LON> = -1.13 +- 5.36" <Off_LAT> = 0.43 +- 5.56" Write SCAN-AVG.dat
The applied corrections are stored in Pointing.dat the data for further processing in LIST.raw and the averages over LON and LAT of the raw scans are written in SCAN-AVG.dat. The LIST.raw file now contains one entry for each scan with the following information.
# JD Scan Source Flux Err Azi. Elv. UT LST par.Ang #------------------------------------------------------------------------------- # 15519.4576 6894 3C454.3 27.5893 0.1693 261.8 27.7 22.98 3.30 41.0 15622.2729 6896 3C48 2.6540 0.0159 271.8 43.7 18.55 5.62 49.4 15622.3106 6905 3C295 2.6072 0.0189 38.4 26.4 19.45 6.53 -40.0 15622.3167 6908 3C295 2.5999 0.0148 39.5 27.3 19.60 6.68 -41.2 15622.3370 6914 3C295 2.6271 0.0156 43.3 30.4 20.09 7.16 -45.2 ....
The next step will be to correct this data for the gain elevation dependence and to convert it from Kelvin to Jansky.
Final Calibration
For frequencies below 2.5 GHz the gain of the Effelsberg antenna is more or less constant over the whole elevation range, but at high frequencies a gain curve should be applied to the data to correct for the losses. The gain curve for each receiver is given on the receiver page. The Fortran-program eff_flux can be used to correct the data for various effects including the gain curve and to calculate and apply the conversion factor from Kelvin to Jansky.
The program requests an input file called "eff_flux.par". It is structured as follows, sorry most of the variable are called in German, but some comments are introduced on the right side here to explain the meaning.
##Control file for eff_flux
ANZAHL QUELLEN: [ 5] # number of sources
##Source name: 8 characters
3C273
3C279
3C286
3C295
3C309.1
3C345
4C39.25
NRAO150
OJ287
##----------------------------------------------
ANZAHL ELEVATIONSKORREKTUREN: [ 1] # number of elevation corrections
##source name ('0836+71') or 'allsour', JD time interval, poly. coeff. A0-A5
'allsour' 0.0 99999.9 0.88196 6.6278E-3 -9.2334E-5 0.0 0.0 0.0
##----------------------------------------------
KORREKTURKURVE (y/n): [n] # is there a free correction curve*
##----------------------------------------------
ANZAHL ZEITKORREKTUREN: [ 2] # number of time correction
##source (s.o.), JD time interval, coeff. A0-A3, and p,B1,B2 with
## B1*sin(p*t) und B2*cos(p*t)
'allsour' 0.0 99999.9 1.000 0.0 0.0 0.0 0.0 0.0 0.0
'NGC7027' 0.0 99999.9 0.981 0.0 0.0 0.0 0.0 0.0 0.0 # constant factor to correct fora partly
# resolve calibrator
##----------------------------------------------
ANZAHL KALIBRATIONEN: [ 1] # number of calibrations
##AJD time interval, cal.-factor,-error
0.0 99999.9 1.0 0.0
##----------------------------------------------
ANZAHL KALIBRATORQUELLEN: [ 8] # number of prime calibrators to calculate K/Jy
## name and flux density in Jy ('3C286' 7.58)
'3C286' 2.82
'3C295' 1.18
'3C196' 0.91
'3C138' 1.57
'3C48' 1.40
'3C161' 1.57
'3C147' 2.07
'3C123' 3.79
* the correction curve should be named "Corr_curve" and has the following form: three columns with
JD corr-fact. err
Some example eff_flux.par files can be found in /home/obs2/flux_monit/eff_flux-par/. The eff_flux program creates several files.
- LIST.corr contains the corrected entries from LIST.raw
- FLUX.<source name> contain the individual calibrated scans of each source
- Averages gives a summary for all soruces
- Calibrators reports the data of prime calibrators and states the conversion factor from Kelvin to Jansky. The factor can then be written into the eff_flux.par. After the value was entered in eff_flux.par the Calibrators file should give 1.0 as the calibrations factor.
The output of the program looks as follows
obs2@observer3:~$ eff_flux **************************************************** * Analysis of flux density * * measurements made at the 100m * * radio-telescope in Effelsberg * * * * Version 2.4 * * Alexander Kraus, MPIfR * * * **************************************************** Files needed: LIST.raw (total-power-Data) and the commandfile Command.tot. Maybe a time-dependent correction (File Corr_curve). Commandfile read! Starts to read data from LIST.raw 100 data sets read! Derives averages!! Starts writing results!!
Done!

